
Second, analzye covariation after recalibration of raw data: Note that we will be using the previously mentioned dbSNP file (dbsnp.vcf) in this step.įirst, analyze covariation patterns in raw data: This is necessary, because the sequencing machine may incorrectly estimate the base quality scores due to various sources of systemic technical error. In this step, we will attempt to recalibrate the per-base quality scores assigned by the sequencing machine. targetIntervals realignment_targets.list \ Please note that we will be using the previously mentioned 1000 Genomes Project Gold Standard Indels file (gold_indels.vcf) to accomplish this.įirst, we mark where realingments should occur in our BAM files: In this step, we will use GATK to remove mapping artifacts that might be mistaken for SNPs. This is the first step to actually use GATK. The output of this command should be a "dedup_reads.bai" file in the same directory as the BAM file. INPUT=aligned_reads_with_readgroupinfo.bam \Īfter this, we must create indexes for the resulting BAM files with: This can be done with Picard using the following command: This should have already been done to produce the starting BAM files. This is a large topic in and of itself, and thus, outside the scope of this guide. The first step in Pre-Processing is "Map to Reference", or in other words, alignment (Click "Map to Reference" in the chart on the right). The solution to this obstacle was found here.

See this link for more info on the Picard commands Next, to sort the BAM file to match the contig order of the reference FastA file, use the following command: OUTPUT=aligned_reads_with_readgroupinfo.bam \ Java -jar picard.jar AddOrReplaceReadGroups \ You can both coordinate sort and add read group info to a BAM file with Picard using the following command: This is critical, since the latest versions of GATK will not proceed if this info isn't present in the BAM files. Let's take a look at what Pre-Processing for RNA-Seq data involves.īefore we can do any of these, however, we need to be sure that our aligned BAM files are coordinate sorted with regard to our reference and contain read group information. The steps of Pre-Processing are different depending on the type of raw data you're starting with. The original documentation for this step can be found here. So without further ado, let's get started. More information on this graphic can be found here.Īs you can see, variant calling is broken into three main subprocesses: Lucky for us however, the Broad Institute has an even prettier graphic outlining the whole thing: Unfortunately, the process for calling SNPs with GATK isn't quite as simple as the pretty graphic above would lead you to believe. (Optional) To make your life a little easier, a cluster of powerful computers and the proper tools to run things on them.A working copy of Picard (another very useful bioinformatics tool), to iron out minor kinks in the files listed above.A working copy of the Broad Institute's Genome Analysis Toolkit (GATK).Files containing well-known-mutation sites for your organism in VCF format.
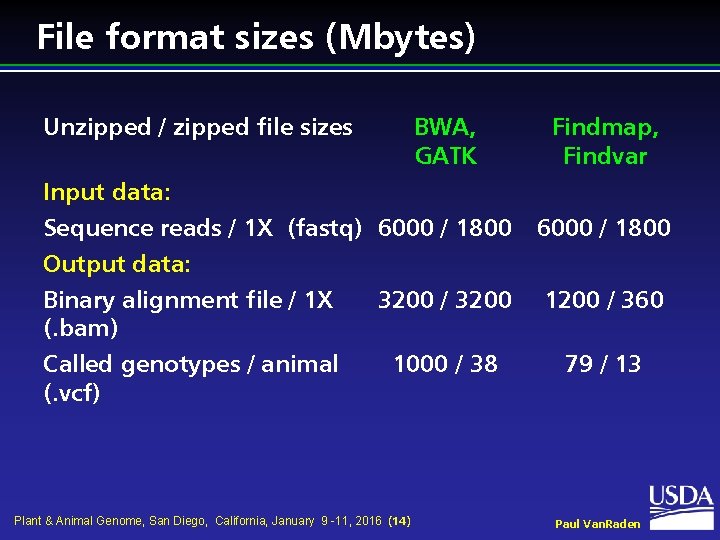
A reference genome for your sample organism (hg19 in my case) in FastA format.The sequenced and aligned genome of your sample organism (human fibroblast cells in my case) in BAM format.More info on the input files to GATK can be found here. A more in-depth explanation of this can be found here.įor my analysis, I used the dbSNP and 1000 Genomes Project Gold Standard Indels known-mutation-sites files (in VCF format). It's one of those powerful features I mentioned earlier that makes GATK so great. I should also mention that GATK can use files containing well-known mutation sites in the sample organisms genome to make its own mutation finding more accurate. GATK was then used to find mutations in the resulting BAM files. The FastQ files were aligned to a reference genome (hg19, in FastA format) to produce BAM files. The sample cells' genomes were gained through whole transcriptome shotgun sequencing (RNA-Seq), which produced raw data files in the FastQ format. Here's a brief summary of what I was trying to do and why I needed GATK to do it:Īs an undergraduate researcher at my universities Systems Biology lab, my goal was to look into whether changes in the physical shape of the genetic material in human fibroblast cells could be linked to certain mutations in the cells' genomes. Using the official GATK documentation as a guide, I will attempt to highlight some of the obstacles I encountered while calling variants with GATK, with the hope that this info might help someone avoid the same pitfalls.Even still, it's easy to run into hitches on the long journey from raw sequencer data to filtered and annotated SNPs.GATK is one of the most powerful and well-documented tools avaliable for calling varaints.


 0 kommentar(er)
0 kommentar(er)
